Papers, Reviews and Book Chapters
(59) Bodin, M. R. and Hammond, M. C. "Visualizing intracellular glycine with two-dye and single-dye ratiometric RNA-based sensors" Nucleic Acids Res (2025) in press.
Related Highlights: Nominated by all reviewers and selected by NAR editors as Breakthrough Article
(58) Ricks, N. J., Brachi, M., McFadden, K., Jadhav, R. G., Minteer, S. D., and Hammond, M.C. “Development of Malate Biosensor-Containing Hydrogels and Living Cell-Based Sensors”
Int. J. Mol. Sci. 2024, 25(20), 11098 Link

(57) Lone, M.S., Merino-Chavez, O. D., Ricks, N. J., Hammond, M.C., Noriega, R. “Electron Transfer Drives the Photosensitized Polymerization of Contrast Agents by Flavoprotein Tags for Correlative Microscopy”
J. Am. Chem. Soc. 2024, 146, 34, 23797–23805 Link
Related Highlights: "In search of new microscopy tools to observe how cells function": U chemists discover how key contrast agent works, paving way to create markers needed for correlative microscopy.

(56) Mumbleau, M.M., Chevance, F., Hughes, K., Hammond, M.C. “Investigating the effect of RNA scaffolds on the multi-color fluorogenic aptamer Pepper in different bacterial species”
ACS Synth. Biol. (2024) 13, 4, 1093–1099 Link

(55) Squire S.*, Sebghati S.*, Hammond, M.C. “Cytoplasmic accumulation and permeability of antibiotics in Gram positive and Gram negative bacteria visualized in real-time via a fluorogenic tagging strategy”
ACS Chem. Biol. (2024) 19, 1, 3–8 Link to paper. Link(Chem Archive) *co-first authors

(54) Manna S., Kimoto, M., Truong, J., Bommisetti, P., Peitz, A., Hirao, I., Hammond, M.C. “Breaking riboswitch speed limits by systematic mutation and orthogonal base pair incorporation”
ACS Sens. (2023) 8, 12, 4468–4472 Link to paper. Link(Chem Archive)

(53) Banuelos Jara, B. and Hammond, M.C. "Functions and applications of riboswitches" Handbook of Chemical Biology of Nucleic Acids" (2022) (Invited Chapter) Link
(52) Mumbleau, M.M., Meyer, M.R., Hammond, M.C. "Determination of In Vitro and Cellular Turn-on Kinetics for Fluorogenic RNA Aptamers" JOVE (2022) J. Vis. Exp. (186), e64367 (Invited Paper) Link
(51) Mukkayyan, N., Poon, R., Sander, P.N., Lai, L-Y., Zubair-Nizami, Z., Hammond, M.C., Chatterjee, S.S. "In Vivo Detection of Cyclic-di-AMP in Staphylococcus aureus" ACS Omega (2022) 7 (36), 32749-32753 Link
(50) Lowry, R.C., Hallberg, Z.F., Till, R., Simons, T.J., Nottingham, R.,Want, F., Sockett, R.E.,Lambert, C., Hammond, M.C. "Production of 3', 3' -cGAMP by a Bdellovibrio bacteriovirus promiscuous GGDEF enzyme, Bd0367, regulates exit from prey by gliding motility" PLoS Genetics (2022) 18(5): e1010164 Link
Related Highlights: Double take: A dual-functional Hypr GGDEF synthesizes both cyclic di-GMP and cyclic GMP—AMP to control predation in Bdellovibrio bacteriovorus PLoS Genetics


(49) Tan, Z., Chan, C.H., Maleska, M., Banuelos Jara, B., Lohman, B.K., Ricks, N.J., Bond, D.R., Hammond, M.C. "The Signaling Pathway That cGAMP Riboswitches Found: Analysis and Application of Riboswitches to Study cGAMP Signaling in Geobacter sulfurreducens" International Journal of Molecular Science, (2022) 23(3), 1183 Link

(48) Manna, S.*, Truong, J.*, Hammond, M.C. "Guanidine biosensors enable comparison of cellular turn-on kinetics of riboswitch-based biosensor and reporter" ACS Synth Biol (2021) 10, 3, 566-578 Link
*co-first authors

(47) Manna, S., Kellenberger, C.A., Hallberg, Z.F., Hammond, M.C. "Live cell imaging using riboswitch-spinach tRNA fusions as metabolite-sensing fluorescent biosensors" Methods Mol Bio (2021) 2323, 121-140 (Invited Chapter) Link
(46) Kitto, R. Z.*, Dhillon, Y.*, Bevington, J.* et. al. Welch, C., McKay, C. P., Hammond, M. C. “Synthetic biological circuit tested in spaceflight” Life Sci Space Res (2021) 28, 57-65 Link
*co-first authors

(45) Palmer, A. E., Hammond, M. C. “Editorial overview: Molecular imaging” Curr Opin Chem Biol (2020) 57, A5-A7 Link
(44) Kitto, R.Z., Christiansen, K.E., Hammond, M.C. "RNA-based fluorescent biosensors for live cell detection of bacterial sRNA" Biopolymers (2020) (Invited Paper) e23394 Link

(43) Yao, L., Fin A., Rovira, A.R., Su, Y., Dippel, A.B., Valderrama, J.A., Riestra, A.M., Nizet, V., Hammond, M.C., Tor, Y. "Tuning the innate immune response to cyclic dinucleotides using atomic mutagenesis" ChemBioChem (2020) 21, 18, 2595-2598 Link
(42) Anderson, W.A., Dippel, A.B., Maiden, M.M., Waters, C., Hammond, M. C. “Chemiluminescent sensors for quantitation of the bacterial second messenger cyclic di-GMP” Methods in Enzymology (2020) 640, 83-104 (Invited Chapter) Link

(41) Su, Y. and Hammond, M. C. “RNA-based fluorescent biosensors for live cell imaging of small molecules and RNAs” Curr Opin Biotech (2020) 63, 157-166 (Invited Review) Link

(40) Dippel, A. B.*, Anderson, W. A.*, Park, J. H., Yildiz, F. H., Hammond, Ming. C. "Development of ratiometric bioluminescent sensors for in vivo detection of bacterial signaling" ACS Chem Biol (2020) 15, 4, 904-914 Link
*co-first authors

(39) Wright, T. A., Jiang, L., Park, J. J., Anderson, W. A., Chen, G., Hallberg, Z. F., Nan, B., and Hammond, M. C. "Second messengers and divergent HD-GYP enzymes regulate 3',3'-cGAMP signaling" Molecular Microbiology (2020) 113, 222-236 Link

(38) Wright, T. A., Dippel, A. B., Hammond, M. C. “Cyclic di-GMP signaling gone astray: cGAMP signaling via Hypr GGDEF and HD-GYP enzymes” In Chou, S.-H., Guilliani, N., Lee, V., Romling U. (ed), Microbial cyclic di-nucleotide signaling. (Invited Chapter) Link
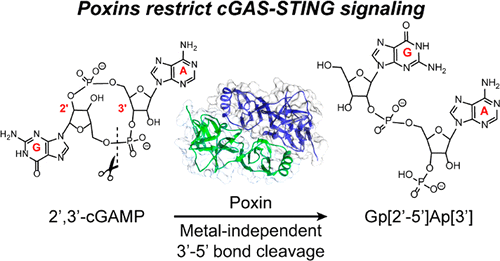
(37) Dippel, A. B. and Hammond, M. C. "A poxin on both of your houses: Poxviruses degrade the immune signal cGAMP" Biochemistry (2019) 58, 19, 2387-2388 (Invited Viewpoint) Link
Head
(36) Hallberg, Z. F.*, Chan, C. H.*, Wright, T. A., Park, J. J., Kranzusch, P. J., Doxzen, K., Bond,
D. R., Hammond, M. C. “Structure and mechanism of a Hypr GGDEF enzyme that activates cGAMP signaling to control extracellular metal respiration” eLife (2019); e43959 Link
*co-first authors
ing 1

Headi
(35) Villa, J.*, Su, Y.*, Contreras, L. M., Hammond, M. C. “Synthetic biology of small RNAs and riboswitches” (2019) 527-545. In Storz G, Papenfort K (ed), Regulating with RNA in Bacteria and Archaea. ASM Press, Washington, DC. doi: 10.1128/microbiolspec.RWR-0007-2017 (Invited Book Chapter) Link
*co-first authors
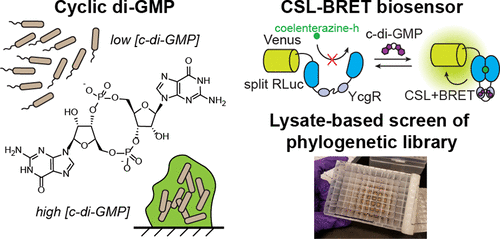
(34) Dippel, A. D., Anderson, W. A., Evans, R. S., Deutsch, S., Hammond, M. C. "Luminescent biosensors for detection of second messenger cyclic di-GMP" ACS Chem Bio (2018) 13, 1872-1879 (Invited Paper) Link
Related Highlights: "Sensors" special issue; Research Highlight in Nat Chem Biol, JGI Science Highlight
(33) Truong, J., Hsieh, Y.-F., Truong, L., Jia, G., Hammond, M.C. "Designing fluorescent biosensors using circular permutations of riboswitches", Methods (2018) 143, 102-109. (Invited Paper) Link
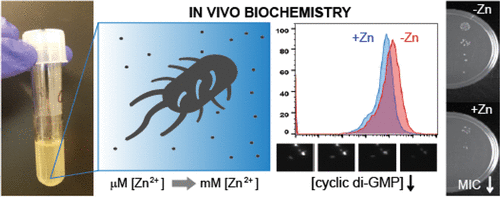
(32) Yeo, J., Dippel, A. B., Wang, X. C., Hammond, M. C. "In Vivo Biochemistry: Single-cell dynamics of cyclic di-GMP in E. coli in response to zinc overload" Biochemistry (2018) 57, 108-116. Link
Related Highlights: Future of Biochemistry special issue; College of Chemistry news story
(31) Yeo, J., Wang, X. C., Hammond, M. C. "Live flow cytometry analysis of c-di-GMP levels in single cell populations" Methods Mol Biol (2017) 1657, 111-130. Link to Book
(30) Hallberg, Z. F., Su, Y., Kitto, R. Z., Hammond, M. C. "Engineering and in vivo applications of riboswitches" Annual Rev Biochem (2017) 86, 515-539. Link
(29) Bose, D.*, Su, Y.*, Marcus, A., Raulet, D. H., Hammond, M. C. "An RNA-based fluorescent biosensor for high-throughput analysis of the cGAS-cGAMP-STING pathway" Cell Chem Biol (2016) 23, 1539-1549.
*co-first authors Link

(28) Wang, X. C., Wilson, S. C., Hammond, M. C. "Next-generation RNA-based fluorescent biosensors enable anaerobic detection of cyclic di-GMP" Nucleic Acids Res (2016) 44, e139. Link
(27) Su, Y., Hickey, S. F., Keyser, S. G. L., Hammond, M. C. "In vitro and in vivo enzyme activity screening via RNA-based fluorescent biosensors for S-adenosyl-L-homocysteine (SAH)" J Am Chem Soc (2016) 138, 7040-7047. Link

(26) Hallberg, Z. F., Wang, X. C., Wright, T. A., Nan, B., Ad, O., Yeo, J., Hammond, M. C. "Hybrid promiscuous (Hypr) GGDEF enzymes produce cyclic AMP-GMP (3', 3'-cGAMP)" Proc Natl Acad Sci (2016) 113, 1790-1795. Link
Related Highlights: 2015 NIH High Risk-High Reward Symposium; Berkeley news story
(25) Muller, R. Y., Hammond, M. C., Rio, D. C., Lee, Y. J. "An efficient method for electroporation of small interfering RNAs (siRNAs) into ENCODE Project Tier 1 GM12878 and K562 cell lines" J Biomol Tech (2015) 26, 142-149. Link
(24) Gonzalez, T. L., Liang, Y., Nguyen, B., Staskawicz, B. J., Loque, D., Hammond, M. C. "Tight regulation of plant immune responses by combining promoter and suicide exon elements" Nucleic Acids Res (2015) 43, 7152-7161. Link
(23) Kellenberger, C. A., Sales-Lee, J., Pan, Y., Gassaway, M. M., Herr, A. E., Hammond, M. C. "A minimalist biosensor: quantitation of cyclic di-GMP using the conformational change of a riboswitch aptamer" RNA Biol (2015) 12, 1189-1197. Link
(22) Kellenberger, C. A.*, Chen, C.*, Whiteley, A. T., Portnoy, D. A., Hammond, M. C. "RNA-based fluorescent biosensors for live cell imaging of second messenger cyclic di-AMP" J Am Chem Soc (2015) 137, 6432-6435. Link
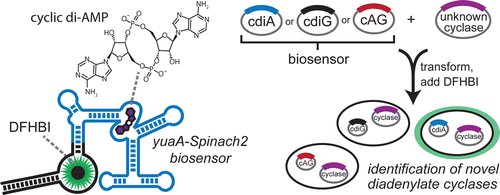
(21) Ren, A., Wang, X. C., Kellenberger, C. A., Rajashankar, J. R., Jones, R., Hammond, M. C., Patel, D. J. "Structural basis for molecular discrimination by a 3', 3'-cGAMP sensing riboswitch" Cell Reports (2015), 11, 1-12. Open Access PDF

(20) Kellenberger, C. A.*, Wilson, S. C.*, Hickey, S. F., Gonzalez, T. L., Su, Y., Hallberg, Z. F., Brewer, T. F., Iavarone, A. T., Carlson, H. K., Hsieh, Y. F., Hammond, M. C. "GEMM-I riboswitches from Geobacter sense the bacterial second messenger c-AMP-GMP" Proc Natl Acad Sci (2015) 112, 5383-5388.
*co-first authors Link
Related Highlights: 2015 Signaling Breakthroughs of the Year
(19) Kellenberger, C. A., Hallberg, Z. F., Hammond, M. C. "Live cell imaging using riboswitch-Spinach tRNA fusions as metabolite-sensing fluorescent biosensors" Methods in Mol Biol (2015) 1316, 87-103. Link to Chapter
(18) Kellenberger, C. A., Hammond, M. C. "In vitro analysis of riboswitch-Spinach aptamer fusions as metabolite-sensing fluorescent biosensors" Methods Enz (2015) 550, 147-172. Link to Chapter
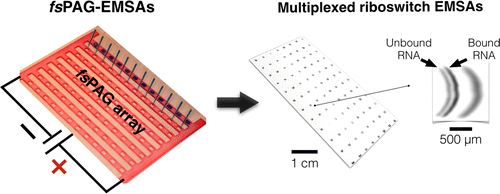
(17) Pan, Y., Duncombe, T. A., Kellenberger, C. A., Hammond, M. C., Herr, A. E. "High-throughput electrophoretic mobility shift assays for quantitative analysis of molecular binding reactions" Analytical Chem (2014) 86, 10357-10364. Link
(16) Wilson, S. C., Cohen, D. T., Wang, X. C., Hammond, M. C. "A neutral pH thermal hydrolysis method for quantification of structured RNAs" RNA (2014) 20, 1153-1160. Link
(15) Hickey, S. F., Hammond, M. C. "Structure-guided design of fluorescent S-adenosylmethionine analogs for a high-throughput screen to target SAM-I riboswitch RNAs" Chem Biol (2014) 21, 345-356. Link

(14) Sadhu, M. J., Guan, Q., Li, F., Sales-Lee, J., Iavarone, A. T., Hammond, M. C., Cande, W. Z., Rine, J."Nutritional control of epigenetic processes in yeast and human cells" Genetics (2013) 195, 831-844. Link
(13) Diner, E. J., Burdette, D. L., Wilson, S. C., Monroe, K. M., Kellenberger, C. A., Hyodo, M., Hayakawa, Y., Hammond, M. C., Vance, R. E. "The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING" Cell Reports (2013), 3, 1355-1361. Link

(12) Leppek, K., Schott, J., Reitter, S., Poetz, F., Hammond, M. C., Stoecklin, G. "Roquin promotes constitutive mRNA decay via a conserved class of stem-loop recognition motifs" Cell (2013) 153, 869-881. Link

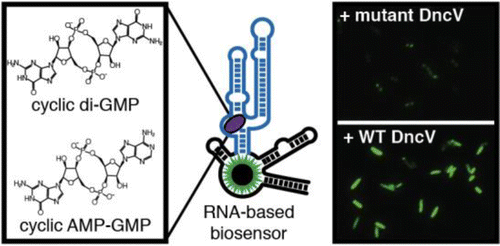
(11) Kellenberger, C. A., Wilson, S. C., Sales-Lee, J., Hammond, M. C. "RNA-based fluorescent biosensors for live cell imaging of second messengers cyclic di-GMP and cyclic AMP-GMP" J Am Chem Soc (2013) 135, 4906-4909. Link
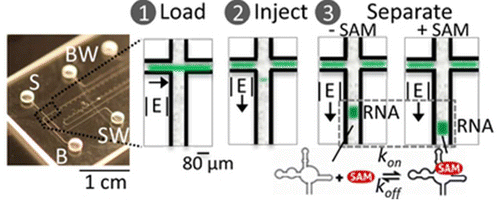
(10) Karns, K., Vogan, J. M., Qin, Q., Hickey, S. F., Wilson, S. C., Hammond, M. C., Herr, A. E."Microfluidic screening of electrophoretic mobility shifts elucidates riboswitch binding function" J Am Chem Soc (2013), 135, 3136-3143. Link
(9) Hickey, S. F., Sridhar, M., Westermann, A. J., Qin, Q., Vijayendra, P., Liou, G., Hammond, M. C."Transgene regulation in plants by alternative splicing of a suicide exon" Nucleic Acids Res (2012), 40, 4701-4710. Link
Selected by NAR editors as a Featured Article
(8) Hammond, M. C. "A tale of two riboswitches" Nat Chem Biol (2011), 7, 342-3. Link
=========================================================
(7) Meyer, M. M., Hammond, M. C., Salinas, Y., Roth, A., Sudarsan, N., Breaker, R. R. "Challenges of ligand identification for riboswitch candidates" RNA Biol (2011), 8, 5-10. Link
(6) Block, K. F., Hammond, M. C., Breaker, R. R. "Evidence for widespread gene control function by the ydaO riboswitch candidate" J Bacter (2010), 192, 3983-9. Link
(5) Hammond, M. C., Wachter, A., Breaker, R. R. "A plant 5S rRNA mimic regulates alternative splicing of transcription factor IIIA pre-mRNAs" Nat Struct and Mol Biol (2009), 16, 541-9. Link
(4) Weinberg, Z., Regulski, E. E., Hammond, M. C., Barrick, J. E., Yao, Z., Ruzzo, W. L., Breaker, R. R. "The aptamer core of SAM-IV riboswitches mimics the ligand-binding site of SAM-I riboswitches" RNA (2008), 14, 822-8. Link
(3) Hammond, M. C., Bartlett, P. A. "Synthesis of amino acid-derived cyclic acyl amidines for use in beta-strand peptidomimetics" J Org Chem (2007), 72, 3104-07. Link
(2) Hammond, M. C., Harris, B. Z.; Lim, W. A., Bartlett, P. A. "Beta-strand peptidomimetics as potent PDZ ligands" Chem Biol (2006), 13, 1247-51. Link
(1) Sudarsan, N.*, Hammond, M. C.*, Block, K. F., Welz, R., Barrick, J. E., Roth, A., Breaker, R. R. "Tandem riboswitch architectures exhibit complex gene control functions" Science (2006), 314, 300-304.
*co-first authors Link
Patents and Patent Applications
(6) Hammond, M. C., Wright, T. A. "Methods of producing cyclic dinucleotides" U.S. Pat Appl Publ 62/438,126 (2016).
(5) Hammond, M. C., Su, Y., Bose, D. "Fluorescent biosensor for 2', 3'-cGAMP" U.S. Pat Appl Publ 62/349,556 (2016).
(4) Hammond, M. C., Su, Y., Keyser, S. G. L., Hickey, S. F. "A fluorescent biosensor for high throughput screening of methyltransferase activity" U.S. Pat Appl Publ 62/246,953 (2015).
(3) Vance, R. E., Hammond, M. C., Burdette, D., Diner, E. J., Wilson, S. C. "Cyclic di-nucleotide induction of type I interferon" U.S. Pat Appl Publ 14/268,967 (2014).
(2) Hammond, M. C., Westerman, A. J., Qin, Q. "A P5SM suicide exon for regulating gene expression" U.S. Pat Appl Publ 13/747,395 (2013).
(1) Bartlett, P. A., Hammond, M. C. "Peptide beta-strand mimics based on pyridinones, pyrazinones, pyridazinones, and triazinones" U.S. Pat Appl Publ (2005).
